> p1 <- ChlorReads(9876, "Virgil", "M", 248, 45, 148) > print(p1) ID: 9876 Name: Virgil Gender: M LDL: 248* HDL: 45+ Triglycerides: 148* means high risk value, + means borderline. Use these cutoff values the three chloresterol values:
| Type | Range | Meaning | Symbol |
|---|---|---|---|
| LDL | 0 to 200 | Normal | "" |
| LDL | 200 to 240 | Borderline | "+" |
| LDL | 240 and up | High Risk | "*" |
| HDL | 0 to 40 | High Risk | "*" |
| HDL | 40 to 60 | Borderline | "+" |
| HDL | 60 and up | Normal | " " |
| Trigl | 0 to 150 | Normal | "" |
| Trigl | 150 to 200 | Borderline | "+" |
| Trigl | 200 and up | High Risk | "*" |
You can use an if-else statement to obtain the symbol. However, be careful not to start a new line with else:
# Correct
if (score >= 90) {
grade = "A"
} else if (score >= 80) {
grade = "B"
} else {
grade = "C"
}
# Incorrect
if (score >= 90) {
grade = "A"
}
else if (score >= 80) {
grade = "B"
}
else {
grade = "C"
}
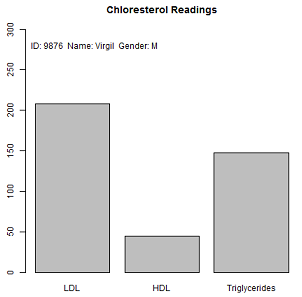
> getPatientInfo(p1) ID Name Gender 9876 Virgil M > getChloresterol(p1) LDL HDL Trigl 208 45 148
- Tests the methods defined in 1 through 4.
- Uses the data in chlor-reads.txt to create a list lst of
ChlorReads objects indexed as
[[1]], [[2]], ... , [[length(lst)]]. To do this, you can use a for loop to process the rows of the
data frame:
lst <- NULL chlor <- read.table("chlor-reads.txt") for(i in 1:nrow(chlor)) { lst[[i]] <- ChlorReads(chlor[i, ]$id, as.character(chlor[i, ]$name), as.character(chlor[i, ]$gender), chlor[i, ]$ldl, chlor[i, ]$hdl, chlor[i, ]$trigl) }The as.character method calls for the name and gender components are because these components are stored as factor objects in a data frame.
- Uses a for loop to print all of the ChlorReads objects
in lst.
- Uses a for loop to plot all of the ChlorReads objects in lst to a PDF file.