2021 DePaul MedIX REU Project Description
· Predict Prognosis of Age-Related Macular Degeneration using OCT Images
· Analysis of CT scans to evaluate normal tissue complications in radiation therapy patients
· Soft segmentation of lung nodules in CT images
· Automatic evaluation of grey-white matter differentiation in CT
· Learning with uncertain labels for CAD
· Human-in-the-Loop similarity retrieval
· Blood protein analysis in patients suffering chronic fatigue
· Hybrid Human-Machine Materials Structure Discovery
· Hybrid Human-Machine Scientific Information Extraction
Predict Prognosis of Age-Related Macular Degeneration using OCT Images
Advanced form of age-related macular degeneration (AMD) is a major health burden that can lead to irreversible vision loss in the elderly population. For early preventative interventions, there is a lack of effective tools to predict the prognosis outcome of advanced AMD because of the similar visual appearance of retinal image scans in the early stage and the variability of prognosis paths among patients. An early characteristic of AMD is drusen, which appears as yellowish deposits under the retina (Figure 1 left). AMD is mainly categorized into two types: Dry AMD (non-neovascular, Figure 1 middle) is represented by drusen deposition, later evolving into confluent areas of regressed drusen and ultimately in the advanced dry stage presenting as loss of vision associated with retinal pigment epithelium (RPE) atrophy (clinically known as geographic atrophy, GA). Wet AMD (neovascular, Figure 1 right) is characterized by the leakage of fluid in the sub-RPE and subretinal spaces caused by neovascularization. The overall objective for this study is to design, develop, and evaluate AMD prognosis prediction models that can detect most relevant images containing AMD biomarkers, manage unevenly spaced sequential optical coherence tomography (OCT) images and predict all advanced AMD forms that can help with the interpretation and explainability of computer-aided prognosis models. Read more
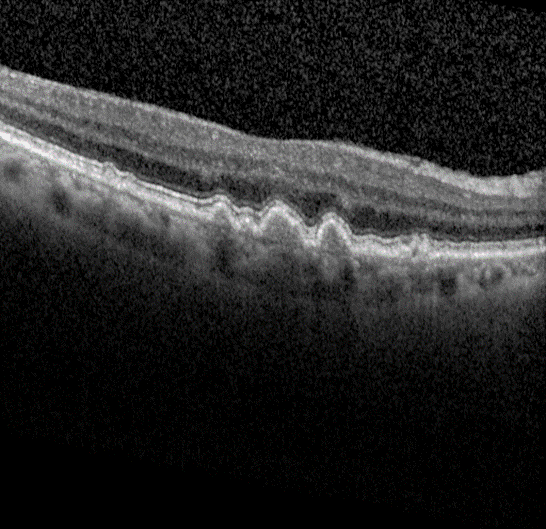
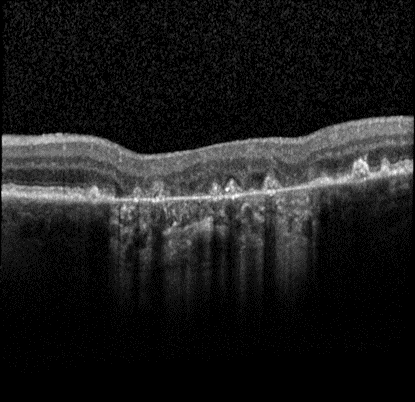
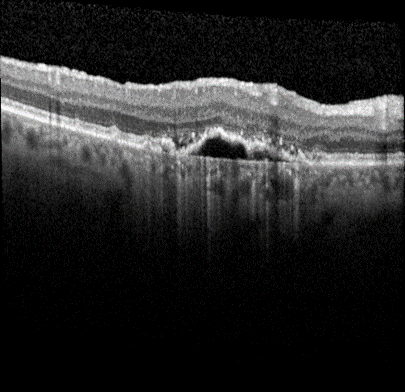
Figure 1: Dursen (left), Dry AMD (middle) and Wet AMD (right)
Analysis of CT scans to evaluate normal tissue complications in radiation therapy patients
Patients who undergo radiation therapy for lung cancer often are monitored for tumor response through post-therapy CT scans. These images may also demonstrate early signs of normal tissue toxicities induced by the radiation. This project investigates computerized image analysis methods to identify, quantify, and analyze potential image-based signs of lung damage due to radiation therapy. Students will work with advanced image registration algorithms to spatially align the pre- and post-therapy CT scans of radiation therapy patients. Read more
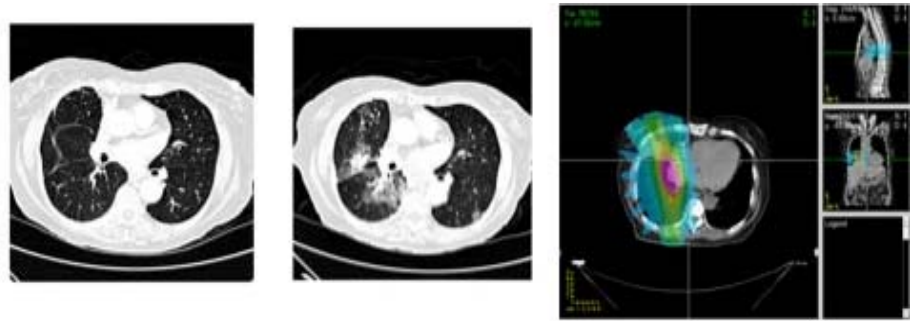
Figure 2: (left) Baseline CT scan from a lung cancer patient undergoing radiation therapy. (center) The same CT section after radiation therapy demonstrating normal lung tissue damage. (right) The radiation dose map from a different patient. The expectation is that regions of increased normal tissue damage correspond with regions of higher radiation dose.
Soft segmentation of lung nodules in CT images
Producing accurate and consistent segmentations of lung nodules in CT scans facilitates the extraction of more accurate quantitative imaging measurements that can be later used for more efficient computer-aided diagnosis (CAD). While there are several “hard segmentation” approaches presented in the literature, there are only a few studies that present “soft segmentation” probabilistic approaches. In this project, students will investigate new machine learning approaches to (1) produce soft segmentation of lung nodules, (2) understand how a set of computer-generated weak segmentations compare with manually generated segmentations in terms of generating qualitative image features for nodule diagnostic classification, and (3) determine the generalizability of their algorithm to other anatomical structures and imaging modalities. Read more
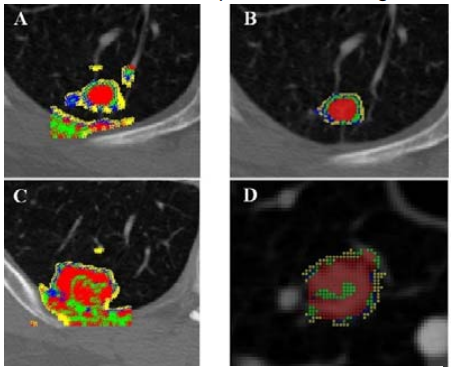
Automatic evaluation of grey-white matter differentiation in CT
Cardiac arrest, with only 12% out-of-hospital and 24.8% in-hospital survival rates, leaves many living patients comatose. Challenges in properly predicting outcomes concerning life span, disability, and neurological damage of patients with return of spontaneous circulation can negatively affect decisions regarding hospital management. As a result, it is necessary to find a proper metric to aid physicians making prognoses. Following various case studies showing that grey matter attenuation of x-rays decreases during ischemia and hypoxia, several research studies showed that clinical outcome can be predicted with high specificity using the ratio of Hounsfield Units (HU) in grey matter to that in white matter, referred to as the grey-white ratio (GWR). Some studies used regions of interest (ROIs) placed around specific tissue types, while others used novel CT segmentation methods for a more global approach. Both approaches provide a highly specific prediction of poor outcomes but have low sensitivity. It is possible that sensitivity could be increased by a method that determines GWRs for not only the entire brain but also for individual lobes. In this REU project, students will develop a pipeline for segmentation of grey and white matter and further compare the distributions of Hounsfield Unit (HU) values in each tissue type. Using the Montreal Neurological Institute (MNI) 152 template and lobe masks in the same standard space to identify the regions of interest on CT scans, students will also develop new approaches for patient CT data normalization to make their algorithms generalizable across patient populations. Read more
Figure 4: Tissue specific decomposition of a native CT to determine GWR in global cerebral hypoxia. Automated probabilistic assessment of gray matter (middle column) and white matter (right column) in computed tomography (left column): GWR’s in two patients resuscitated after cardiac arrest with good outcome versus poor outcome (lower row). This image comes from the paper by Hanning et. al.
Learning with uncertain labels for CAD
Creating classifiers for CAD in the absence of ground truth is a challenging problem. Using experts’ opinions as a reference truth is difficult because the variability in the experts’ interpretations introduces uncertainty in the labeled diagnostic data. This uncertainty translates into noise, which can significantly affect the performance of any classifier on test data. To address this problem, students will investigate (1) novel approaches to combine experts’ interpretations, (2) probabilistic classification approaches to deal with uncertainty, and (3) conformal prediction approaches to provide confidence and credibility in the CAD outcome. Read more
Figure 5: Visual representation of the LIDC data structure; one nodule is exemplified through the differences in the nodule’s outlines and semantic ratings.
Human-in-the-Loop similarity retrieval
Image retrieval, the identification of images similar to a query image, enables physicians to draw upon previous cases to improve diagnosis. However, retrieving images based on their content remains a challenging problem given the semantic gap between the image features automatically extracted from raw pixel data and the high-level interpretation of image content. Convolutional and Siamese neural networks have recently been shown to produce excellent results in image classification and similarity retrieval. These networks are composed of multiple layers that, rather than segmenting an image, process small portions of an input image to learn the relevant features. Capitalizing on these machine learning advances and using the NIH/NCI LIDC data, students will 1) evaluate the retrieval performance of Siamese neural networks, 2) compare them with other more traditional feature engineering methods such as Gabor filters and wavelet transforms as well as with the human visual perception of similarity, and 3) finally integrate the human feedback into an iterative approach for image similarity learning and refinement. The traditional approaches of this project have been successfully advanced by REU students during the previous and will serve as reference literature and reference results for new REU participants. Read more
Blood protein analysis in patients suffering chronic fatigue
Chronic fatigue syndrome is a prevalent disease with little known about its probable cause. Some research has shown a link to the Epstein-Barr virus, implicated in mononucleosis. In this project, we will perform complex statistical analysis of a data set gathered by the DePaul Psychology department; a data set of blood proteins measured in students at three stages: healthy volunteers, students who developed mono and a six month follow-up. We will analyze individual proteins in an attempt to be able to predict whether individuals are likely to develop chronic fatigues syndrome after getting mono, and also use correlational analysis to determine if there are patterns of protein co-activation that characterize healthy controls differently than individuals with chronic fatigue syndrome. A truly multi-disciplinary project, this project will also include regular meetings with the research group at the Psychology department.
Hybrid end-to-end learners
One of the most powerful tools in the computer vision toolbox is the convolutional neural net: a combination of learned multi-scale convolutions and a fully connected deep neural net. Unfortunately, while able to tackle an immense variety of complex vision tasks, the fully connected deep neural net is a black box: there are few satisfying ways to characterize in intuitive or easily explained ways how the neural classifier makes its decisions. On the other hand, the deep neural nets provide a seamless mechanism to learn weights from a given loss function all the way back to the initial weights of the multi-scale convolutions. This project will explore replacing the fully connected deep neural net with more traditional classifiers, such as decision trees, which are explainable and intuitive. This project will require significant knowledge of differential calculus.
Hybrid Human-Machine Materials Structure Discovery
The efficient discovery of new materials can provide crucial solution to current challenges in sustainability, e.g., more efficient fuel cell technology. One of the ways materials scientists discover new materials is through the exploration of ternary (composed of three materials) samples of alloys. They study and interpret high-intensity X-ray diffraction patterns of material spread onto a silicon wafer. Scientists schedule experiments using costly instruments at special facilities such as the Cornell High Energy Synchrotron Source at Cornell University. Access to such instrument is also limited to a few days/weeks a year. Hence, decreasing the time spent during analysis of phase maps would allow researchers to efficiently use the time period scheduled for experimentation, accelerate the discovery process and have significant impact on society. REU students working on this project will investigate a variety of techniques to incorporate domain knowledge and expert input in the clustering of diffraction patterns in order to identify phases in materials sample (samples with the same atom structure).
Hybrid Human-Machine Scientific Information Extraction
Extracting scientific facts from unstructured text is difficult due to challenges specific to the ambiguity of the language, the complexity of the scientific named entities and relations to be extracted. Challenges include acronyms, synonyms, complicated naming conventions, the scarcity of entities in publications (it is not unusual to publish articles about a single newly discovered or synthesized material in polymer science for example). While there exist domain-specific machine learning toolkits that address these challenges, perhaps the greatest challenge is the lack of—time-consuming, error-prone and costly—labeled data to train these machine learning models. REU students working on this project will investigate efficient techniques to involve domain experts and accelerate the process of collecting training data and enabling machine learning techniques to be applied towards scientific information extraction (IE). They will also explore ways to adapt current Natural Language Processing software in order to address the specific challenges of scientific IE. Read more