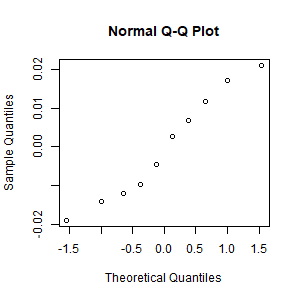
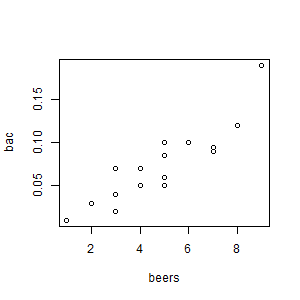- Create the scatter plot of bac vs. beers.
> setwd("c:/it223/beer-bac")
> df1 <- read.csv("beer-bac.txt")
> print(df1)
beers bac
1 5 0.100
2 2 0.030
3 9 0.190
4 8 0.120
5 3 0.040
6 7 0.095
7 3 0.070
8 5 0.060
9 3 0.020
10 5 0.050
11 4 0.070
12 6 0.100
13 5 0.085
14 7 0.090
15 1 0.010
16 4 0.050
Scatterplot of original beer-bac data:
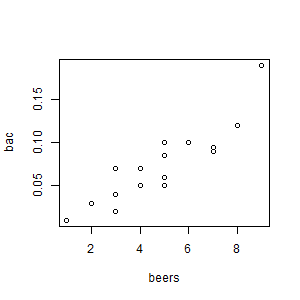
- Find the linear regression equation for predicting bac from beers:
> model1 <- lm(bac ~ beers, data=df1)
> print(model1)
Call:
lm(formula = bac ~ beers, data = df1)
Coefficients:
(Intercept) beers
-0.01270 0.01796
> print(summary(model1))
Call:
lm(formula = bac ~ beers, data = df1)
Residuals:
Min 1Q Median 3Q Max
-0.027118 -0.017350 0.001773 0.008623 0.041027
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.012701 0.012638 -1.005 0.332
beers 0.017964 0.002402 7.480 2.97e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.02044 on 14 degrees of freedom
Multiple R-squared: 0.7998, Adjusted R-squared: 0.7855
F-statistic: 55.94 on 1 and 14 DF, p-value: 2.969e-06
- Find the R-squared value for this equation. Interpret it.
Answer: The r-squared value is 0.7998, which is the proportion of
the variation in the dependent variable due to the variation of
the independent variable. This is a good value for chemistry/biology data.
- Create the boxplot of the residuals:
> residuals <- resid(model1)
> boxplot(residuals)
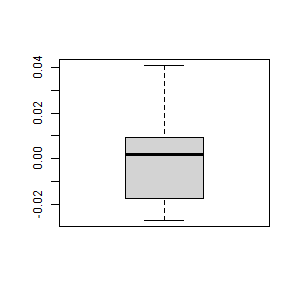
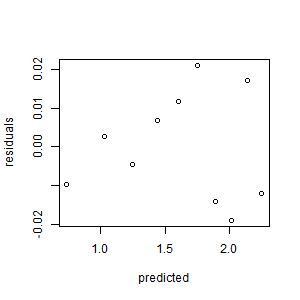
- Create the scatterplot of the residuals vs. the predicted values. Interpret it.
> predicted <- predict(df1)
> residuals <- resid(df1)
> plot(predicted, residuals)
The residuals are unbiased and homoscedastic.
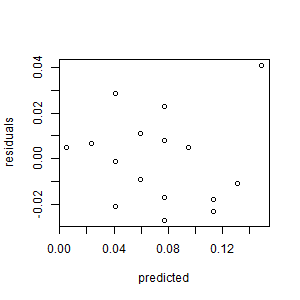
- Create the normal plot of the residuals. Interpret it.
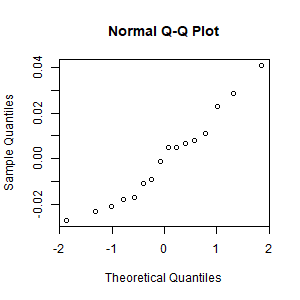
The residuals are
approximately normally distributed.
- For the regression studied in this example, if the number of beers
consumed is 4, what is the predicted blood alcohol level?