Projects
The proposed projects are:
Binning Strategies for Texture Analysis in Computed Tomography (CT) studies.
Wavelets-based methods for texture characterization in Computed Tomography (CT) studies.
Project 1 (Raicu): Binning Strategies for Texture Analysis in Computed Tomography (CT) studies. This project will provide the student researchers an opportunity to understand and develop an interest in the area of image processing and analysis. A texture analysis tool for tissue characterization in CT studies has been developed in our laboratory; the tool uses two statistical texture models (co-occurrence and run-length) and has been already applied to texture characterization of the heart, liver, and renal and splenic parenchyma. The current implementation of the tool deals with DICOM (Digital Imaging and Communications in Medicine) images for which each pixel value is typically contained in the range of 0 to 4096. The current implementation of the texture analysis tool uses 256 bins, each containing 16 gray levels. While there has been some research conducted on binning strategies, they were conducted on images with 256 levels of intensity. Student researchers on this project will investigate the effect of different binning sizes over 4096 intensity levels on differentiating among different types of tissues.
Project 2 (Dettori): Wavelets-based methods for texture characterization in Computed Tomography (CT) studies. Similar to project 1, this project will provide the students an opportunity to understand and develop an interest in the area of image processing and analysis while experimenting with data from the medical domain in the form of DICOM images at different levels of granularity. The texture analysis tool for organ tissues characterization in CT studies will be extended to include other texture models necessary to capture additional properties of texture in the DICOM images. In order to capture the local properties of the texture, student researchers will investigate several wavelets-based methods, such as Gabor filters and curvelets, with respect to the tissues already segmented in the CT images.
Project 3 (Furst): Volumetric texture. This project will allow students to experiment with a large collection of medical CT images and give them first hand experience of interdisciplinary research. The CT data that has been analyzed to date has been taken from multi-detector CT scanners with 4 detectors. It has been, until now, treated as a stack of two-dimensional slices and each image (slice) independently analyzed for texture. Northwestern will be receiving a new 64 slice multi-detector CT scanner in calendar year 2004. With this new technology the CT volume acquired is close to isotropic and will lend itself to the development of volumetric texture characterization as an extension to the texture work already proposed.
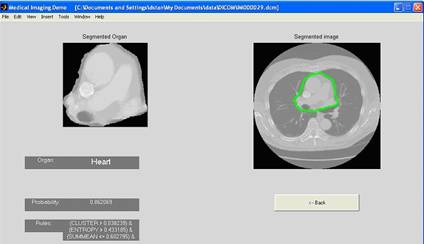
Project 4 (Raicu): Tissue Classification in CT studies. This project will provide the students an opportunity to understand and develop an interest in the area of artificial intelligence, data mining and knowledge discovery. Using the texture analysis tool, a tissue classification tool for the heart, liver, and renal and splenic parenchyma in CT studies has been developed in the IMP laboratory. The main applicability of the tool is to automatically classify tissues in CT images with the goal of early detection of any abnormalities present in these tissues, while increasing the efficiency of radiologists in interpreting the CT images. A screenshot of the existing classification tool is shown in Figure 3. Student researchers will be involved in measuring the performance of the current classifier on new CT images provided by the Northwestern University Department of Radiology and developing new data mining and artificial intelligence strategies (such as neural networks and Bayesian networks) to improve the tissue classification accuracy.

Figure 3: An example of heart segmentation and classification in a DICOM image using co-occurrence and run-length encoding texture descriptors and decision trees
Project 5 (Furst): Tissue Segmentation. Through a rich literature review, student researchers working on this project will understand how texture-based segmentation tools will advance the state of medical image segmentation and increase the efficiency of imaging service operations. The long-term goal of this project is to build an automatic texture-based segmentation tool for normal tissues in CT images. To obtain this objective, the classification rules provided by the classification tool will be used to split and merge different regions in the CT image until a certain probability for texture similarity is met. Further, the vertices of the regions found in the previous step will form the initial points for an active contour algorithm, the most used segmentation algorithm in medical imaging. The expected outcome will be an automatic texture-based segmentation algorithm for healthy tissues in CT images.
Project 6 (Channin): Development of Context Sensitive Reporting Tools. Students will learn about the importance of reporting tools for medical applications. Given segmentation and characterization tools, students will use them to develop structured reporting tools that are context sensitive. Currently, CT studies are interpreted using free-text voice dictation. There is inherently significant variance in how studies are reported. The free text nature of the report makes data mining difficult. This project proposes to develop structured data entry tools that use the RadLex and SNOMED-CT nomenclatures in a DICOM Structured Reporting (SR) context. These tools will be context sensitive as to the tissue identified by the segmentation and classification tools described above. The end result will be direct image annotations of pertinent positive and negative image features using a standardized lexicon and a standardized delivery mechanism. The latter will be based on the Integrating the Healthcare Enterprise Reporting Workflow Integration Profile.
Project 7 (Channin): Performance Analysis. Student researchers will learn to evaluate, interpret and extract knowledge from the results obtained using computer vision and data mining techniques. The above image processing and annotation tools will be integrated into a computer workstation that can be used by radiologists to display and interpret computed tomography (CT) studies. The workstation will query a work-list, retrieve image objects related to a work-list item, provide two- and three-dimensional image display and navigation tools. All of this functionality will be performed in a fashion compliant with the appropriate integration profiles of the Integrating the Healthcare Enterprise (IHE) initiative. Students will then perform receiver operating characteristic studies to demonstrate the impact of the image processing and annotation tools on the accuracy and efficiency of radiologists in this digital environment.
![]()